Publication
Breaking Down Cell-Free DNA Fragmentation: A Markov Model Approach
Terence H.L. Tsui, Phil F. Xie, Salvador Chulián, Víctor M. Pérez-García
biorxiv (doi:10.1101/2023.07.06.547953)
MOLAB authors
Abstract
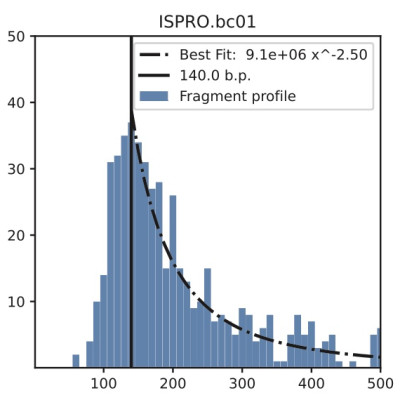
Cell-free DNA (cfDNA) is released into the bloodstream from cells in various physiological and pathological conditions. The use of cfDNA as a non-invasive biomarker has attracted attention, but the dynamics of cfDNA fragmentation in the bloodstream are not well understood and have not been previously studied computationally.
To address this issue, we present a Markovian model called FRIME (Fragmentation, Immigration, and Exit) that captures three leading mechanisms governing cfDNA fragmentation in the bloodstream. The FRIME model enables the simulation of cfDNA fragment profiles by sampling from the stationary distribution of FRIME processes. By varying the parameters of our model, we generate fragment profiles similar to those observed in liquid biopsies and provide insight into the underlying biological mechanisms driving the fragmentation dynamics. To validate our model, we compare simulated FRIME profiles with mitochondrial and genomic cfDNA fragment profiles. Our simulation results are consistent with experimental and clinical observations and highlight potential physicochemical differences between mitochondrial and genomic cfDNA. The FRIME simulation framework provides an initial step towards an improved computational understanding of DNA fragmentation dynamics in the bloodstream and may aid in the analysis of liquid biopsy data.